Enzymatic CarboRelease kit
References:
1. Scudder,P., Uemura, K., Doby, J., Fukuda, M.N. & Feizi, T.(1983) Isolation and characterization of an endo-b-galactosidase from Bacteroides fragilis Biochem. J. 213 , 485-494.
2. Scudder,P., Hanfland, Pl, Uemura, K. & Feizi, T. (1984) Endo-b-galactosidases of Bacteroides fragilis and Escherichia freundii hydrolyze linear but not branched oligosaccharide domains of glycolipids of the neolacto series. J. Biol. Chem . 259, 6586-6592.
3. Scudder, P. Tang, P.W., Hounsell, E.F., Lawson, A.M., Mehmet, H. & Feizi, T. (1986) Isolation and characterization of sulfated oligosaccharides released from bovine corneal keratan sulphate by the action of endo-b-galactosidase. Eur. J. Biochem. 157, 365-373.
4. Murata, T., Hattori, T. Amarume, S. Koicki, A. & Usui, T. (2003) Eur. J. Biochem 270, 3709-3719.
5. Hokke, C.H., Bergwerff, A.A., Van Dedem, D.W., Kamerling, J.P, and Vliegenthart, J.F. (1995) Structural analysis of the N- and O-linked carbohydrate chains of recombinant human erythropoietin expressed in Chinese hamster overay cells. Sialylation patters and branch location of dimeric N-acetyllactosamine units. Eur. J. Biochem. 228, 981-1008.
This kit includes the enzymes and buffers required to deglycosylate glycoproteins, removing all N-linked oligosaccharides and many O-linked sugars.
The enzymes are packaged separately in individual 20 microlitre vials. This gives researchers the flexibility to characterize the glycans attached to their glycoprotein more fully than with our mixture of enzymes, KE-DGMX.
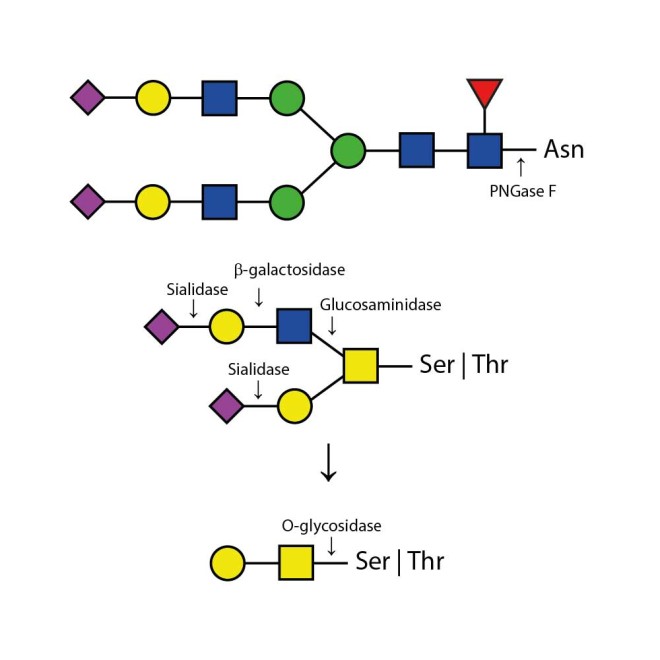
20 µL of each of the following enzymes in separate vials: PNGase F (Elizabethkingia meningosepticum), O-Glycosidase (Streptococcus pneumoniae), Sialidase (Arthrobacter ureafaciens), β-Galactosidase (Streptococcus pneumoniae), Glucosaminidase (Streptococcus pneumonia).
Kit includes enzyme, reaction buffers and Bovine Fetuin (control). Each enzyme is sufficient for up to 20 reactions.
Product specification
Appropriate use of individual glycosidases allow the determination of sialyation (use of Sialidase only) and the specific presence of N-linked glycans (using PNGase F only) and O-linked glycans (using Sialidase, β-Galactosidase, O-Glycosidase, and Glucosaminidase). Alternatively, QA-Bio produces a premixed solution of the enzymes in one 20 microlitre vial in our DeGlycoMx Kit (KE-DGMX).
Appropriate use of individual glycosidases allow the determination of sialyation (use of Sialidase only) and the specific presence of N-linked glycans (using PNGase F only) and O-linked glycans (using Sialidase, β-Galactosidase, O-Glycosidase, and Glucosaminidase). Alternatively, QA-Bio produces a premixed solution of the enzymes in one 20 microlitre vial in our DeGlycoMx Kit (KE-DGMX).
Contents:

Includes 20 µL of each of the following enzymes in separate vials: PNGase F (Elizabethkingia meningosepticum), O-Glycosidase (Streptococcus pneumoniae), Sialidase (Arthrobacter ureafaciens), β-Galactosidase (Streptococcus pneumoniae), Glucosaminidase (Streptococcus pneumoniae).
Also includes:
5x Reaction buffer - 200 µL
Denaturation Solution - 100 µL
Triton X - 100 µL
Bovine Fetuin (control) - 10 mg/mL
Specificity:
The Enzymatic CarboRelease Kit will deglycosylate glycoproteins, removing all N-linked oligosaccharides and many O-linked oligosaccharides from glycoproteins. N-links (Asparganine-linked) are removed using the enzyme PNGase F. In addition, all Serine or Threonine linked (O-linked) Gal-(β1-3)-GalNAc-(α1) and all sialic acid substituted Gal-(β1-3)-GalNAc-(α1) will be removed using the combination of Sialidase and O-Glycosidase. The addition of β-Galactosidase and Glucosaminidase will assist in the deglycosylation of larger O-link structures.
Kit Capacity:
The quantity of enzymes recommended in the protocols is sufficient to deglycosylate approximately 100 µg of an average glycoprotein in the time given. PNGase F cleavage is generally the rate limiting reaction due to the slow removal of some sterically hindered N- linked residues, even when the glycoprotein is denatured. Since all of the enzymes retain activity under reaction conditions for several days, a much larger quantity of glycoprotein may be deglycosylated if incubation is extended. Conversely, there is no need to use the recommended amounts of enzymes if quantities much less than 100 µg of glycoprotein are being cleaved. The enzymes can be diluted into 1X Reaction Buffer. They will remain stable in diluted form at 4°C.
Bovine Fetuin Control Protein:
Bovine Fetuin contains sialylated N- and O- linked oligosaccharides.
NOTE: Commercial preparations of Fetuin contain proteases which will eventually degrade the protein. The Fetuin Control has been heat treated at 90°C for 10 minutes to inactivate the proteases. The Fetuin solution can be stored at 4°C.
Denaturing Protocol:
1. Dissolve 100 µg or less of a glycoprotein in 30 µL deionized water in an Eppendorf tube.
2. Add 10 µL 5X Reaction Buffer and 2.5 µL Denaturation Solution. Mix gently.
3. Heat at 100°C for 5 minutes.
NOTE: Some proteins may precipitate when heated with SDS. In this event, omit the heat treatment and increase the incubation time to 24 hours after adding enzymes.
4. Cool to room temperature. Add 2.5 µL Triton X-100 solution. Mix gently.
NOTE: Failure to add Triton X-100 may result in the reduction of activity of some enzymes.
5. Add 1 µL each of each enzyme.
6. Incubate for 3 hours at 37°C.
7. Analyze by method of choice.
Alternatively, the enzymes may be added individually or sequentially in order to determine what types of oligosaccharides are present on the glycoprotein.
Non-denaturing Protocol:
1. Dissolve 100 µg or less of a glycoprotein in 35 µL deionized water in an Eppendorf tube.
2. Add 10 µL 5X Reaction Buffer.
3. Add 1 µL of each enzyme.
4. Incubate for 1-5 days at 37°C.
An aliquot should be deglycosylated using the denaturing protocol to provide a gel standard for the fully deglycosylated protein. The position of the native protein can then be compared with this standard to judge the extent of deglycosylation.