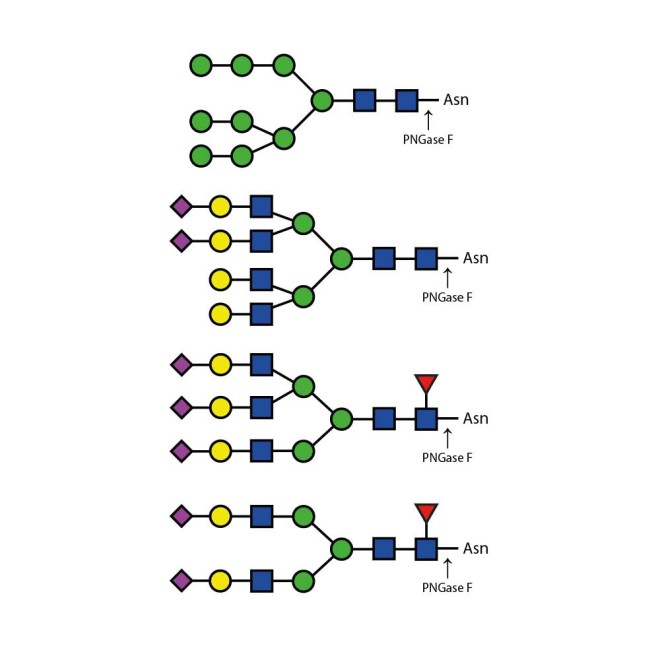
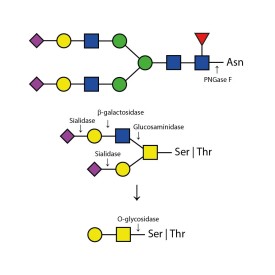
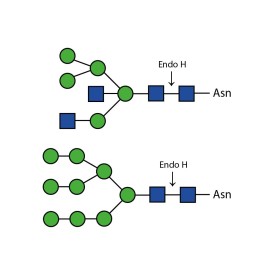
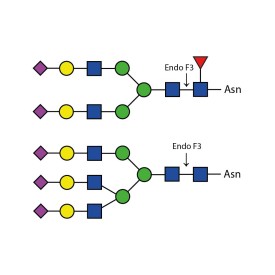
Recombinant PNGase F
References:
1. Bayer, E.A., F. De Meester, T. Kulik and M. Wilchek. Preparation of deglycosylated egg white avidin. Appl Biochem Biotech 53: 1-9 (1995)
2. Elder, J.H. and S. Alexander. endo-b-N-Acetylglucosaminidase F: endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proc Natl Acad Sci USA 79: 4540-4544 (1982)
3. Tarentino, A .L., C.M. Gomez and T.H. Plummer, Jr. Deglycosylation of asparagine-linked glycans by peptide :N-glycosidase F. Biochemistry 24: 4665-4671 (1985)
4. Tarentino A.L. and T.H. Plummer. Enzymatic deglycosylation of asparagine -linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Meth Enzymol 230: 44-57 (1994)
5. Trimble R.B. and A.L. Tarentino. Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1 , endo F2 and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J Biol Chem 266: 1646-1 651 (1991)
6. Taga, E. M., A. Waheed and R. L. Van Etten. Structural and chemical characterization of a homogeneous peptide-N-glycosidase from almond. Biochemistry 23: 815-22 (1984)
7. Tarentino AL, Trimble RB, Plummer TH. Enzymatic approaches for studying the structure, synthesis, and processing of glycoproteins. Methods in Cell Biology: 32: 111-39 (1989)
8. Anthony L. , Tarentino and Thomas H. Plummer Jr. Enzymatic deglycosylation of asparagine-linked glycans: Purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods in Enzymology: 230: 44-57. (1994)
9. Tarentino AL, Plummer TH. Oligosaccharide accessibility to peptide:N-glycosidase as promoted by protein-unfolding reagents. The Journal of Biological Chemistry. 257 (18): 10776-80. (1982)
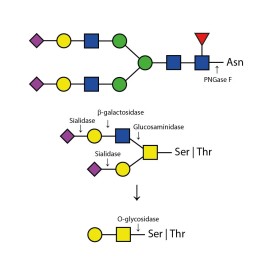
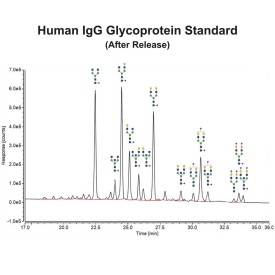
PNGase F is suitable for the release of all types (high-mannose, hybrid and complex) N-linked glycans from glycoproteins and glycopeptides. PNGase F will not remove oligosaccharides containing α(1-3) linked core fucose commonly found on plant glycoproteins.
PNGase F cleaves asparagine-linked (N-linked) oligosaccharides from glycoproteins. PNGase F deaminates asparagine to aspartic acid, leaving the oligosaccharides intact. Denaturation increases the rate of cleavage up to 100x. Most native proteins can still be completely N-deglycosylated but incubation time must be increased. PNGase F will remain active under incubation conditions for at least 72 hours. PNGase F will not remove oligosaccharides containing α(1,3)-linked core fucose commonly found on plant glycoproteins; for this purpose, use peptide N-glycosidase A.
Kit includes enzyme plus reaction buffers. Sufficient for up to 60 reactions.
Product specification:
PNGase F Source: recombinant PNGase F gene from Elizabethkingia miricola in E. coli
EC: 3.5.1.52
Alternative Names: PNGase F, Peptide N Glycosidase F, N-Glycosidase, N-Glycanase
Contents:
60 µL aliquot of recombinant PNGase F (0.3 U) in 20 mM Tris-HCl, pH 7.5
5x PNGase F Reaction Buffer 7.5 for PNGase F - 250 mM sodium phosphate, pH 7.5
PNGase F Denaturation Solution - 2% SDS, 1 M Beta-mercaptoethanol
PNGase F Triton X-100 - 15% solution
Specific Activity: >25 U/mg
Activity: 5 U/mL
Molecular weight: 36,000 daltons
pH range for PNGaseF: 6-10, optimum 7.5
(Protocol) PNGaseF suggested usage:
1. Add up to 200 µg of glycoprotein to an Eppendorf tube. Adjust to 35 µL final volume with de-ionized water.
2. Add 10 µL 5x PNGase F Reaction Buffer 7.5 and 2.5 µL of PNGase F Denaturation Solution. Heat at 100°C for 5 minutes.
3. Cool. Add 2.5 µL of PNGaseF Triton X-100 and mix.
4. Add 2.0 µL of PNGaseF to the reaction. Incubate 3 hours at 37°C.
Specificity: PNGase F cleaves asparagine-linked (N-linked) oligosaccharides from glycoproteins. PNGase F deaminates asparagine to aspartic acid, leaving the oligosaccharides intact. Denaturation increases the rate of cleavage up to 100x. Most native proteins can still be completely N-deglycosylated but incubation time must be increased. PNGase F will remain active under incubation conditions for at least 72 hours. PNGase F will not remove oligosaccharides containing α(1,3)-linked core fucose commonly found on plant glycoproteins; for this purpose, use peptide N-glycosidase A.
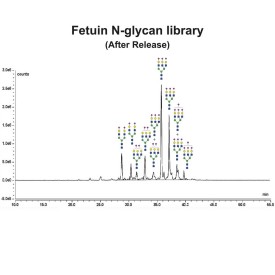
Specific Activity: Defined as the amount of enzyme required to catalyze the release of N-linked oligosaccharides from 1 micromole of denatured RNase B in 1 minute at 37°C, pH 7.5. Cleavage is monitored by SDS-PAGE (cleaved RNase B migrates faster).
Storage: Store enzyme at 4°C.