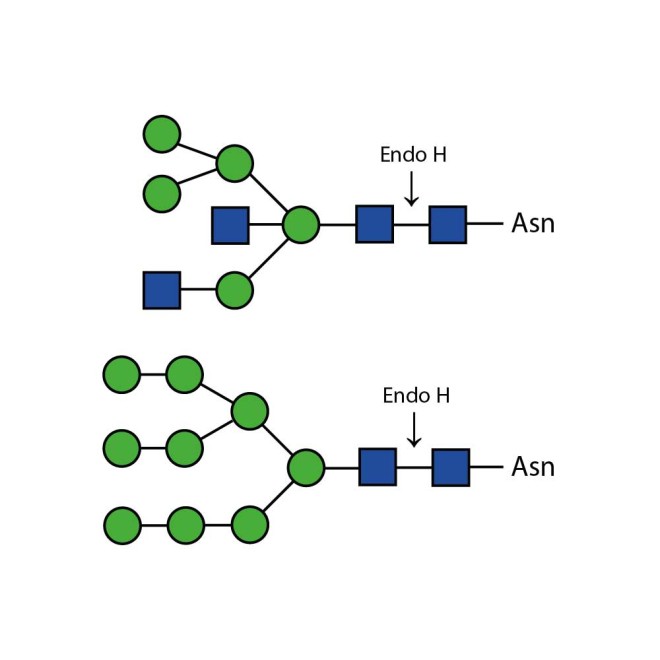
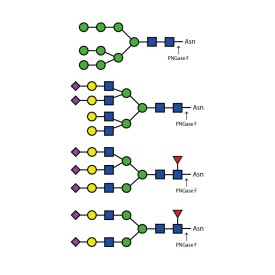
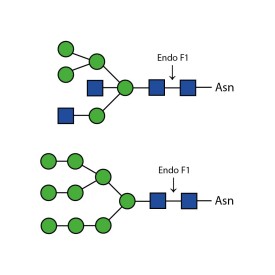
Endoglycosidase H
References:
1. Robbins P. W., D. F. Wirth and C. J. Hering. Expression of the Streptomyces enzyme endoglycosidase H in Escherichia coli. Biol Chem 256:10640-10644 (1981).
2. Robbins P. W., R. B. Trimble, D. F. Wirth, C. Hering, F. Maley , G. F. Maley, R. Das, B. W. Gibson, N. Royal and K. Biemann. Primary structure of the Streptomyces enzyme endo-beta-N-acetylglucosaminidase H. J Biol Chem 259:7577-7583 (1984).
3. Trimble R. B., A. L. Tarentino , G. E Aumick and F. Maley. Endo-beta-N-acetylglucosaminidase L from Streptomyces plicatus. Methods Enzymol 83:603-610 (1982).
4. Trimble R. B. and F. Maley. Optimizing hydrolysis of N-linked high-mannose oligosaccharides by endo-beta-N-acetylglucosaminidase H. Anal Biochem 141:515-522 (1984).
5. Trimble R. B., R. J. Trumbly and F. Maley. Endo-beta-N-acetylglucosaminidase H from Streptomyces plicatus. Methods Enzymol 138:763-770 (1987).
6. Trumbly R. J., P. W. Robbins, M. Belfort, F. D. Ziegler, F. Maley and R. B. Trimble. Amplified expression of Streptomyces endo-beta-N-acetylglucosaminidase H in Escherichia coli and characterization of the enzyme product. J Biol Chem 260:5683-5690 (1985).
Endo H cleaves Asparagine-linked hybrid or high mannose oligosaccharides, but not complex oligosaccharides. It cleaves between the two N-acetylglucosamine residues in the diacetyl chitobiose core of the oligosaccharide, generating a truncated sugar molecule with one N-acetylglucosamine residue remaining on the asparagine. In contrast, PNGase F removes the oligosaccharide intact. Detergent and heat denaturation may increase the rate of cleavage for some glycoproteins.
Kit includes enzyme plus reaction buffers. Sufficient for up to 60 reactions.
Product Specifications:
Source: recombinant from Streptomyces plicatus in E.Coli
EC: 3.2.1.96
Alternative Names: Endo H, endo-β-N-acetylglucosaminidase H, Endoglycosidase H
Specific Activity: One unit of Endo H activity is defined as the amount of enzyme required to catalyze the release of N-linked oligosaccharides from 1 µmole of denatured Ribonuclease B. Cleavage is monitored by SDS-PAGE (cleaved Ribonuclease B migrates faster).
Contents:
60 µL aliquot of enzyme (300 mU) in 20 mM Tris-HCl, 25 mM NaCl, 1 mM EDTA (pH 7.5).
200 µL 5x Reaction Buffer 5.5 (250 mM sodium phosphate, pH 5.5)
Denaturation Solution - 2% SDS, 1 M Beta-mercaptoethanol
Specific Activity: >40 U/mg
Activity: >5 U/mL
Molecular weight: 29,000 daltons
pH range: 5-6, optimum 5.5
Suggested usage:
1. Add up to 200 µg of glycoprotein to Eppendorf tube. Adjust to 37.5 µL final volume with deionized water.
2. Add 10 µL 5X Endo H Buffer and 2.5 µL of Denaturation Solution (SDS/-ME). Heat at 100°C for 5 minutes.
3. Cool, and then add 2.0 µL of Endo H to the reaction. Incubate 3 hours at 37°C.
Storage: Store enzyme at 4°C.
Stability: Stable at least 12 months when stored properly. Several days exposure to ambient tempertures will not reduce activity.
Purity: Endoglycosidase H is tested for contaminating protease as follows; 10 µg of denatured BSA is incubated for 24 hours at 37°C with 2 µL of enzyme. SDS-PAGE analysis of the treated BSA shows no evidence of degradation.
The production host strain has been extensively tested and does not produce any detectable glycosidases.